PROTEOME DATABASE
Naturally occurring urinary peptide and protein database for clinical applications
Urine is an attractive source for biomarker development. It has multiple advantages over other body fluids. Its availability, easy collection and correlation with pathophysiology of a disease are some of the preferable features. Indeed, the possibility of multiple non-invasive sampling and high stability of the peptides are the key factors for state-of-the-art biomarker discovery. However, many proteomic studies have had only limited clinical impact, due to factors such as modest numbers of subjects, absence of disease controls, small numbers of defined biomarkers, and diversity of analytical platforms. CE-MS is an analytical tool that can combine a reasonable analysis time with high resolution, thereby enabling the profiling of urinary samples and recognition of sufficient features to yield robust diagnostic and prognostic panels. CE-MS exhibits excellent performance for biomarker discovery and allows subsequent biomarker sequencing independent of the separation platform. This approach may elucidate the molecular pathogenesis of many diseases and can be used to monitor disease activity and drug response, ultimately improving personalized medicine.
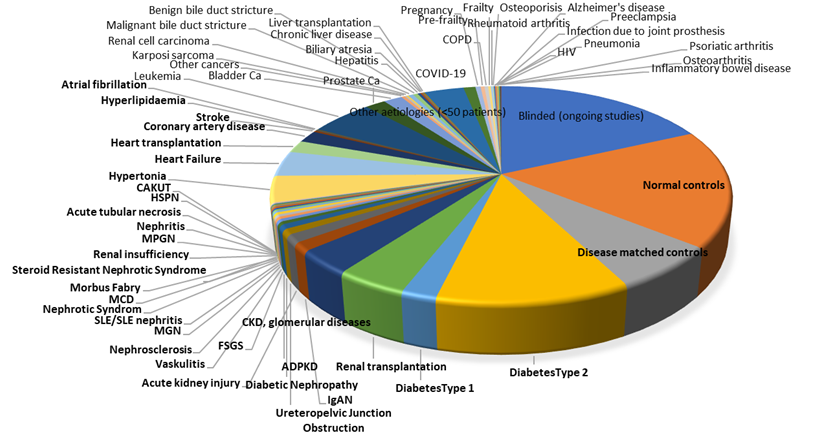
Up-to-date, Mosaiques has generated the largest urinary peptidome/proteome database (molecular range 0.8-20 kDa), currently consisting of more than 85 000 entries from various research, clinical and university centers. This enabled us to define over 20 000 features from which for more than 5000 sequence information was retrieved.
REFERENCES:
Latosinska et al. Electrophoresis. 2019; 40(18-19): 2294-2308
Stallmach et al. Electrophoresis 2013; 34 (11): 1452-1464
Siwy et al. Proteomics Clin. Appl. 2011, 5 (5-6): 367-374
Good et al. Moll Cell Proteomics 2010, 9 (11): 2424-37
Coon et al. Proteomics Clin. Appl. 2008, 2 (7-8): 964
Zürbig et al. Electrophoresis 2006; 27 (11): 2111-2125
For details, please contact:
Prof. Dr. Dr. Harald Mischak
Email: mischak@mosaiques-diagnostics.com
or
Dr. Petra Zürbig
Email: zuerbig@mosaiques-diagnostics.com



