Drug Discovery
Our mission to transform traditional drug discovery

-
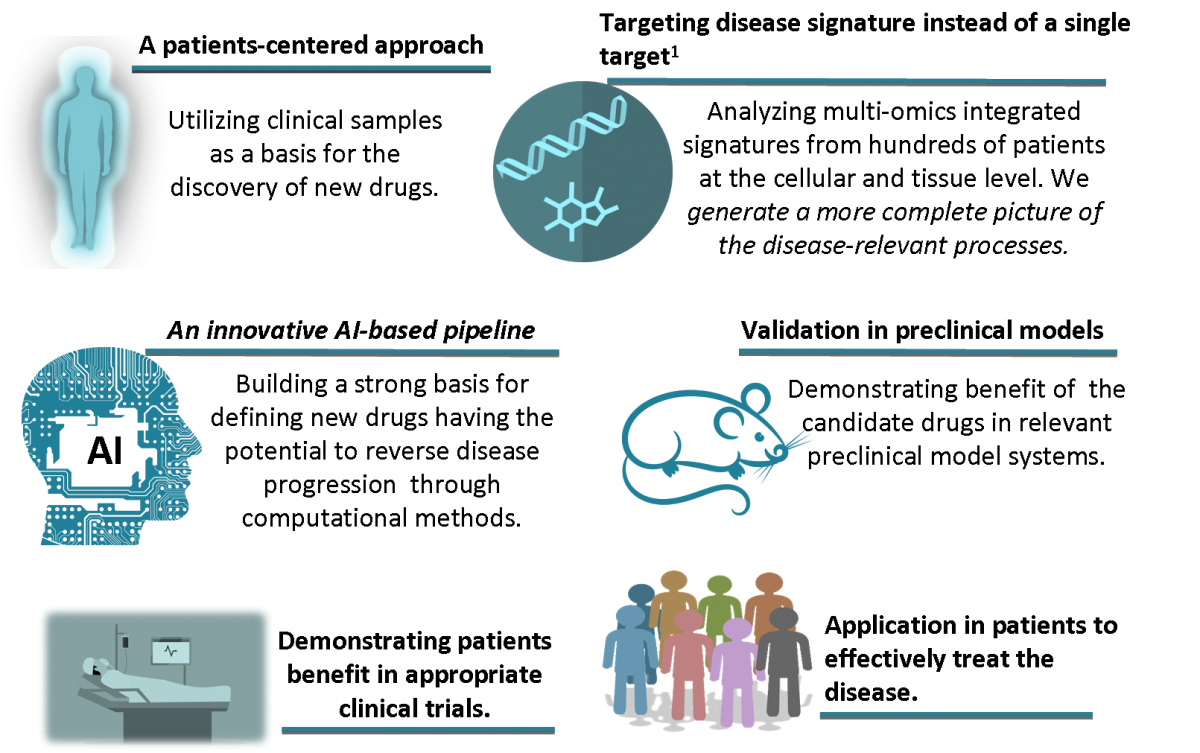
We developed an approach that will radically transform and accelerate drug development and implementation in a personalized manner.
-
This is accomplished trough an innovative pipeline that utilizes an artificial intelligence (AI) based approach.
-
Our pipeline builds on scientific and technological breakthroughs and AI algorithms to integrate high resolution multi-omics data. By this we enable rational selection of drug candidates based on underlying pathophysiology with emphasis on drugs already approved for different context-of-use.
We are committed in delivering novel treatments in oncology
Our approach
-
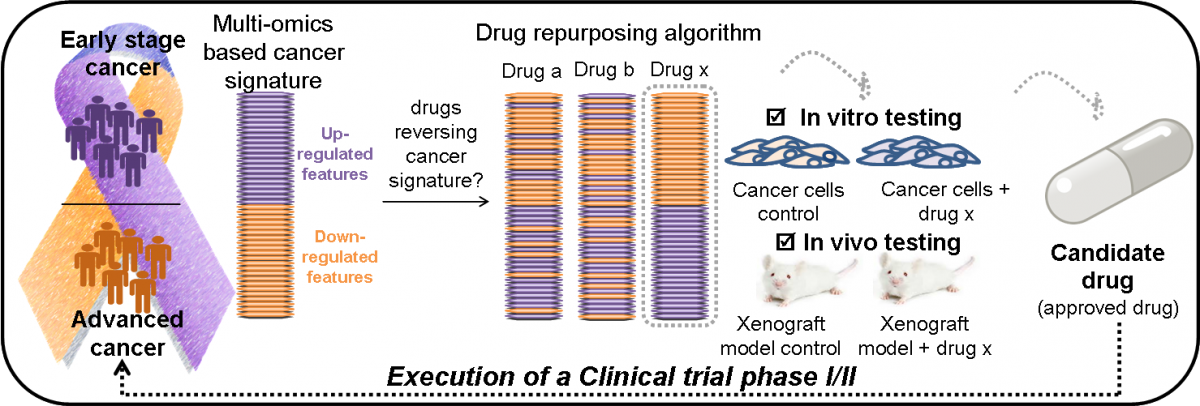
Molecular characterization of cancer progression though integration of high-throughput multi-omics data from patients via advanced computational tools (development of disease signature).
-
Application of an innovative AI-based pipeline to predict drugs having the potential to reverse disease progression.
-
Rationale selection of drug candidates in line with disease pathophysiology.
-
Testing of the candidate drugs in relevant preclinical model systems.
A completely novel all-in-one solution
Exemplifying our approach in bladder cancer (BC)
Generation of BC signature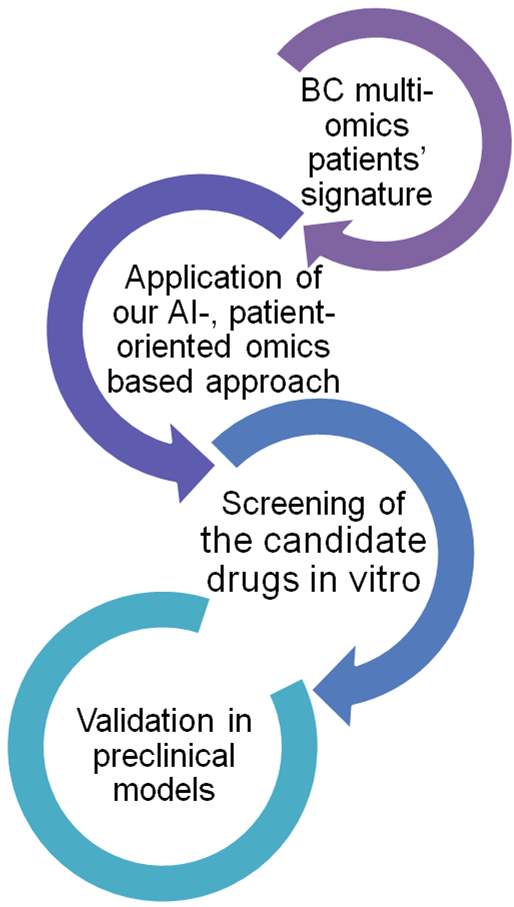
Different -omic signatures were created2 using tissue proteomics3, transcriptomics4 from our previously published works. These were further integrated with omics data retrieved from the literature, but also with in house proteomics and available in repositories transcriptomics data from BC cell lines.
Application of our approach
Using as input in our established drug repurposing pipeline5 these molecular BC signatures , 13 compounds with potential to reverse BC aggressiveness were identified.
In vitro screening
Three of the 13 compounds (namely exemestane, amiodarone and fluvoxamine) demonstrated a prominent impact on cell growth at relatively low concentrations when tested in vitro.
Evaluation in-vivo
These three drugs also showed excellent efficacy in preclinical (animal) models and the next step now is testing these compounds in clinical trials to treat patients with advanced BC that have practically no more treatment option available.
Our specific aim
T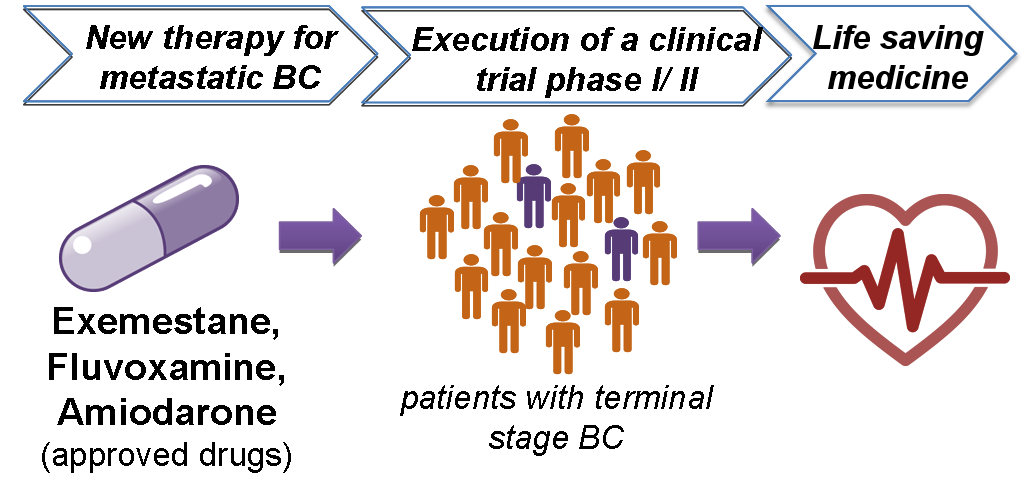 o improve care for BC patients increasing their life expectancy and quality of life
o improve care for BC patients increasing their life expectancy and quality of life
as a result of the novel drugs we introduce.
The next step towards implementation is the execution of a clinical trial phase I/II
We have the full support from the patient organizations (like World Bladder Cancer Patient Coalition) and the relevant professional members (urologists and oncologists) from the European societies of Urology.
Dissemination of the approach during a panel discussion organized by European Medicines Agency (EMA)
https://www.ema.europa.eu/en/events/act-eu-multi-stakeholder-platform-kick-workshop
This work has been supported by
 ReDrugBC is a Marie Sklodowska Curie Actions (MSCA)
ReDrugBC is a Marie Sklodowska Curie Actions (MSCA)
Individual Fellowship programme
funded by the European Commission
(H2020-MSCA-IF-2019, Grant agreement ID: 898260).
Eurostars - Project E! 113726 REDIRECT
CO-FUNDED BY EUREKA MEMBER COUNTRIES AND THE EUROPEAN UNION
HORIZON 2020 FRAMEWORK PROGRAMME
REFERENCES
- Frantzi M. et al. Drug repurposing in oncology. Lancet Oncol. 2020, 21(12):e543. doi: 10.1016/S1470-2045(20)30610-0.
- Mokou M. et al. A Drug Repurposing Pipeline Based on Bladder Cancer Integrated Proteotranscriptomics Signatures. Methods Mol Biol. 2023, 2684:59-99. doi: 10.1007/978-1-0716-3291-8_4
- Stroggilos R. et al. Proteome-based classification of Nonmuscle Invasive Bladder Cancer. Int J Cancer. 2020, 146(1):281-294. doi: 10.1002/ijc.32556.
- Stroggilos R. et al. Gene Expression Monotonicity across Bladder Cancer Stages Informs on the Molecular Pathogenesis and Identifies a Prognostic Eight-Gene Signature. Cancers (Basel). 2022, 14(10):2542. doi: 10.3390/cancers14102542.
- Mokou M. et al. A Novel Pipeline for Drug Repurposing for Bladder Cancer Based on Patients' Omics Signatures. Cancers (Basel). 2020, 12(12):3519. doi: 10.3390/cancers12123519.



